(Para ver la lista completa de publicaciones, da click aqui
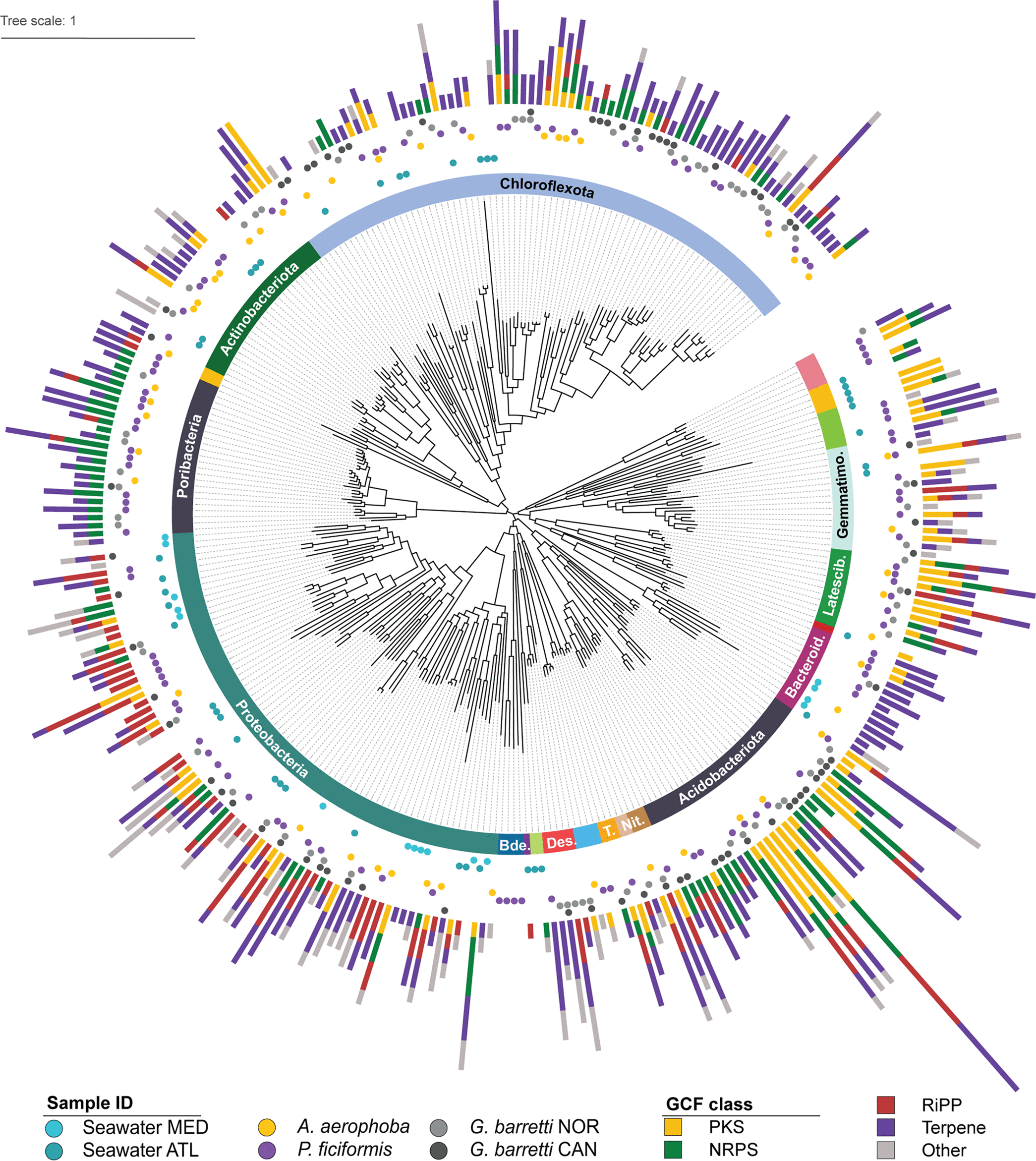
Marine sponges and their microbial symbiotic communities are rich sources of diverse natural products (NPs) that often display biological activity, yet little is known about the global distribution of NPs and the symbionts that produce them. Since the majority of sponge symbionts remain uncultured, it is a challenge to characterize their NP biosynthetic pathways, assess their prevalence within the holobiont, and measure the diversity of NP biosynthetic gene clusters (BGCs) across sponge taxa and environments. Here, we explore the microbial biosynthetic landscapes of three high-microbial-abundance (HMA) sponges from the Atlantic Ocean and the Mediterranean Sea. This data set reveals striking novelty, with <1% of the recovered gene cluster families (GCFs) showing similarity to any characterized BGC. When zooming in on the microbial communities of each sponge, we observed higher variability of specialized metabolic and taxonomic profiles between sponge species than within species. Nonetheless, we identified conservation of GCFs, with 20% of sponge GCFs being shared between at least two sponge species and a GCF core comprised of 6% of GCFs shared across all species. Within this functional core, we identified a set of widespread and diverse GCFs encoding nonribosomal peptide synthetases that are potentially involved in the production of diversified ether lipids, as well as GCFs putatively encoding the production of highly modified proteusins. The present work contributes to the small, yet growing body of data characterizing NP landscapes of marine sponge symbionts and to the cryptic biosynthetic potential contained in this environmental niche.
Catarina Loureiro, Anastasia Galani, Asimenia Gavriilidou, etc.
Research Article, Facultad de ciencias de la Universidad Nacional Sede Bogotá, (2022)
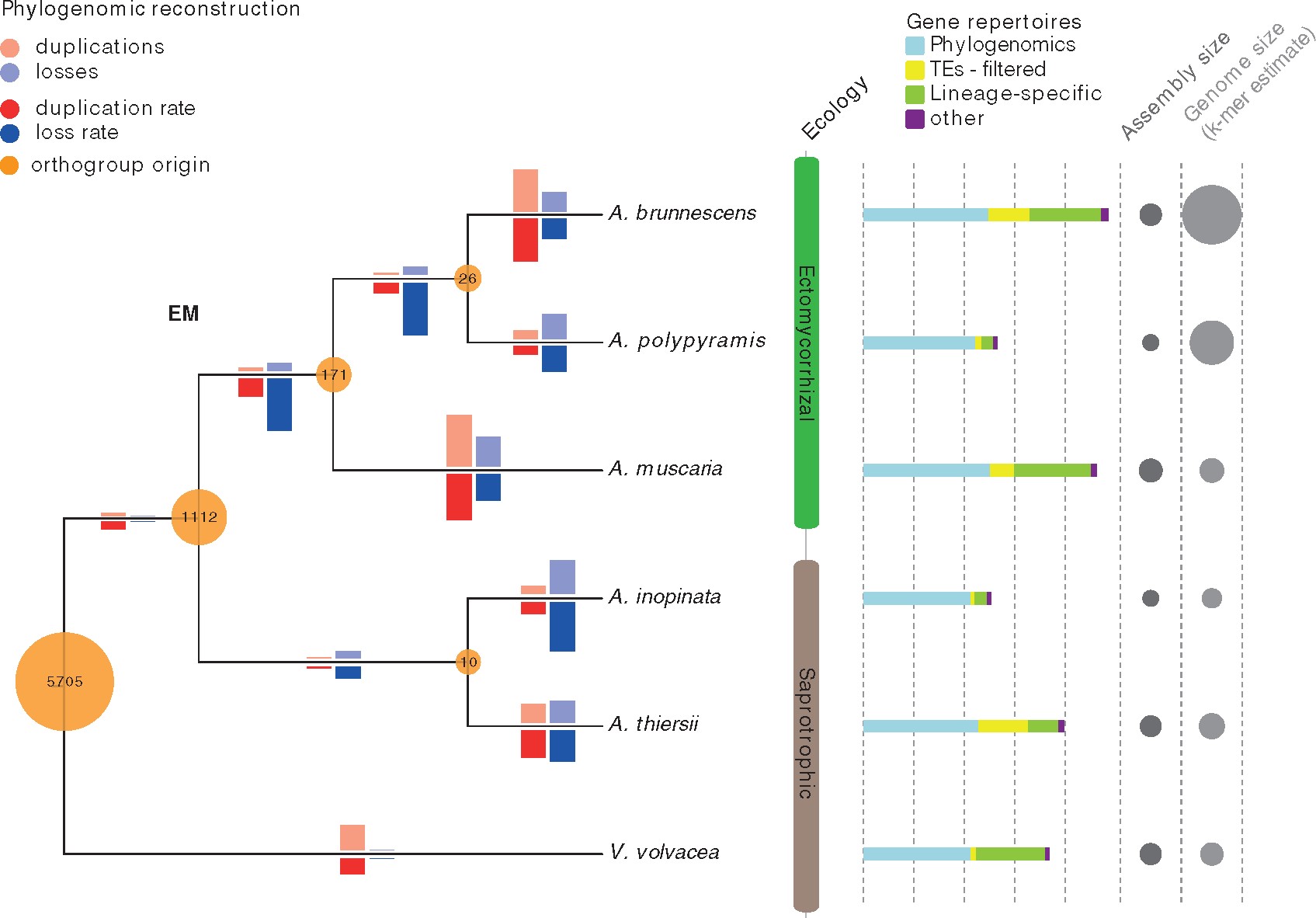
Fungi are evolutionary shape shifters and adapt quickly to new environments. Ectomycorrhizal (EM) symbioses are mutualistic associations between fungi and plants and have evolved repeatedly and independently across the fungal tree of life, suggesting lineages frequently reconfigure genome content to take advantage of open ecological niches. To date analyses of genomic mechanisms facilitating EM symbioses have involved comparisons of distantly related species, but here, we use the genomes of three EM and two asymbiotic (AS) fungi from the genus Amanita as well as an AS outgroup to study genome evolution following a single origin of symbiosis. Our aim was to identify the defining features of EM genomes, but our analyses suggest no clear differentiation of genome size, gene repertoire size, or transposable element content between EM and AS species.
Jaqueline Hess, Inger Skrede, Maryam Chaib De Mares, Matthieu Hainaut, Bernard Henrissat, Anne Pringle
Research Article, Facultad de ciencias de la Universidad Nacional Sede Bogotá, (2022)
@article{GRL5_Mares_Chaib_2025,
title = {Distinct bacteria display genus and species-specific associations with mycobionts in paramo lichens in Colombia},
journal = {Oxford Academic},
author = {Mares, Maryam Chaib De and Castro, Emerson Arciniegas and Ulloa, Maria Alejandra and Torres, Jean Marc and Sierra, Maria A and Butler, Daniel J and Mason, Christopher E and Zambrano, María Mercedes and Moncada, Bibiana and Muñoz, Alejandro Reyes},
year = {2025},
doi = {10.1093/femsec/fiaf010},
image_filename = {},
published = {1},
tag = {10001}
}@article{GRL1_Mares_Chaib2018,
title = {Expressed protein profile of a Tectomicrobium and other microbial symbionts in the marine sponge Aplysina aerophoba as evidenced by metaproteomics},
journal = {Nature},
author = {Mares, Maryam Chaib De and Jiménez, Diego Javier and Palladino, Giorgia and Gutleben, Johanna and Lebrun, Laura A. and Muller, Emilie E. L. and Wilmes, Paul and Sipkema, Detmer and van Elsas, Jan Dirk},
year = {2018},
doi = {10.1038/s41598-018-30134-0},
image_filename = {./images/publications/GRL1_Mares_Chaib2018.webp},
published = {1},
tag = {10001}
}@article{GRL2_Langenberger_David2009,
title = {Evidence for human microRNA-offset RNAs in small RNA sequencing data},
journal = {Oxford University Press},
author = {Langenberger, David and Bermudez-Santana, Clara and Hertel, Jana and Hoffmann, Steve and Khaitovich, Philipp and Stadler, Peter F.},
year = {2009},
doi = {10.1093/bioinformatics/btp419},
image_filename = {./images/publications/GRL2_Langenberger_David2009.png},
published = {1},
tag = {10002}
}@article{GRL4_Mares_Chaib2023,
title = {Design of gene circuits in microorganisms to modulate host physiology},
journal = {Sociedad Colombiana de Ciencias Químicas},
author = {Gomez, Edgar Babativa and Naranjo, Julieth Camila and Loaiza, Luis Miguel Riveros and Vega, Catalina and Medina, Juan Carlos Riveros and Gomez, Francisco},
year = {2023},
image_filename = {./images/publications/GRL4_Mares_Chaib2023.png},
published = {1},
tag = {10003}
}@article{GRL3_Mares_Chaib2014,
title = {Horizontal transfer of carbohydrate metabolism genes into ectomycorrhizal Amanita},
journal = {New Phytologist Foundation},
author = {Mares, Maryam Chaib De and Hess, Jaqueline and Floudas, Dimitrios and Lipzen, Anna and Choi, Cindy and Kennedy, Megan and Grigoriev, Igor V. and Pringle, Anne},
year = {2014},
doi = {10.1111/nph.13140},
link = {https://nph.onlinelibrary.wiley.com/doi/10.1111/nph.13140},
image_filename = {./images/publications/GRL3_Mares_Chaib2014.webp},
published = {1},
tag = {10004}
}@article{GRL6_Mares_Chaib2018,
title = {Rapid Divergence of Genome Architectures Following the Origin of an Ectomycorrhizal Symbiosis in the Genus Amanita},
journal = {Oxford Academic},
author = {Hess, Jaqueline and Skrede, Inger and Mares, Maryam Chaib De and Hainaut, Matthieu and Henrissat, Bernard and Pringle, Anne},
year = {2018},
doi = {10.1093/molbev/msy179},
link = {https://academic.oup.com/mbe/article/35/11/2786/5100885},
image_filename = {},
published = {1},
tag = {10004}
}@article{GRL7_Mares_Chaib2022,
title = {Functional and Phylogenetic Characterization of Bacteria in Bovine Rumen Using Fractionation of Ruminal Fluid},
journal = {frontiers},
author = {Hernández, Ruth and Mares, Maryam Chaib De and Jimenez, Hugo and Reyes, Alejandro and Caro-Quintero, Alejandro},
year = {2022},
doi = {10.3389/fmicb.2022.813002},
link = {https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2022.813002/full},
image_filename = {},
published = {1},
tag = {10004}
}